Feature Importance
Ben Jakubowski
NYU
May 1st, 2018
Learning objectives¶
Feature Importance¶
- (Informally) define feature importance, and explain why exploring and presenting feature importance can be both (i) useful, and (ii) potentially misleading.
- Define and describe several feature importance methods that exploit the structure of the learning algorithm or learned prediction function.
- Describe a prediction-function-agnostic method for generating feature importance scores.
- Describe the limitations of these feature importance measures and understand cases where they "fail".
Partial Dependence Plots¶
- Describe how partial dependence plots are generated.
- Describe the limitations of these plots, and some potential remedies.
Defining feature importance¶
- Problem setup -- we have some data $(x, y) \in (\mathbb{R}^{d}, \mathcal{Y})$, and a learned prediction function $f$.
- A feature importance method can be loosely understood as a function that maps each feature onto some score.
- These scores rank features by how much they "contribute" to the prediction function $f$.
- Here "contribute" is defined separately for each method.
- In general, feature importance is not consistently or rigorously defined.
Why we need to discuss feature importance¶
- It is easy to compute feature importance from many ML libraries.
Why we need to discuss feature importance¶
- It is easy to compute feature importance from many ML libraries.
- This means feature importance bar charts show up all over, and it is important to understand:
- How feature importance can be computed.
- Potential pitfalls in over interpreting these scores.
Utility of feature importance scores¶
- Question: Why might feature importance scores be useful?
- Use for sanity checking a model -- do the features that are important seem reasonable, or do they suggest something strange is going on under the hood (i.e. leakage)?
- Share with clients/bosses to build trust and get buy-in on the prediction function.
- Use for feature selection -- can extract the top $K$ features and retrain over this subset (ex:
sklearn.feature_selection.SelectFromModel)
Feature selection example¶
- Breiman's original Random Forests paper showed feature importance for classification of political party based on 16 votes[1](https://link.springer.com/content/pdf/10.1023/A:1010933404324.pdf):
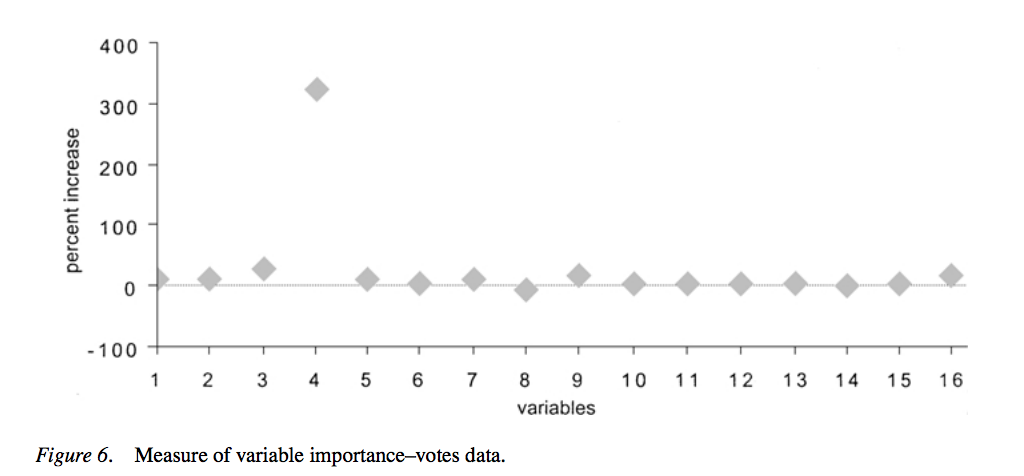
- 'We reran this data set using only variable 4. The test set error is 4.3%, about the same as if all variables were used.'
First example/introducing the issues¶
- Consider the following feature importance plot for a regression problem with $$\mathcal{Y} = \mathcal{A} = \mathbb{R}, X = \mathbb{R}^{11}$$
hide_code_in_slideshow()
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
from sklearn.linear_model import ElasticNetCV
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
sns.set_context('notebook', font_scale=1.5)
%matplotlib inline
X,y = make_regression(n_samples=1000, n_features=10,
n_informative=3,random_state=1234,
shuffle=False, noise=10)
X = np.append(X, X[:,[0]], axis=1)
train_X, test_X, train_y, test_y = train_test_split(X,y,
test_size=0.2,
random_state=1235)
enet = ElasticNetCV(l1_ratio=np.array([.1, .5, .7, .9, .95, .99, 1]),
n_alphas=500,
normalize=True,
selection='random',
random_state=1351)
enet.fit(train_X, train_y)
coefs = pd.DataFrame({'Feature': np.arange(len(enet.coef_)),
'Importance': np.abs(enet.coef_)})
coefs = coefs.set_index('Feature')\
.sort_values('Importance', ascending=False)
fig, ax=plt.subplots(figsize=(12,6))
plot = coefs.plot(kind='bar', title='Example feature importance chart', ax=ax)
- Question: What can we say about this prediction function, it features, and target?
First example/introducing the issues (continued)¶
- Recall the full feature set includes features $\{0,\cdots,10\}$.
- Let's compare both (a) test set performance ($R^2$), and (b) feature importance plots, for models trained on:
- All features
- Features $\{0,\cdots,9\}$
- Features $\{1,\cdots,9\}$
- Features $\{1,\cdots,10\}$
First example/introducing the issues (continued)¶
- Recall the full feature set includes features $\{0,\cdots,10\}$...
from matplotlib import rc
rc('text', usetex=True)
from sklearn.metrics import mean_squared_error
from math import floor
hide_code_in_slideshow()
features_to_test = {
'All': np.arange(0,11),
'0-9': np.arange(0,10),
'1-9': np.arange(1,10),
'1-10': np.arange(1,11)
}
fig, axarr = plt.subplots(ncols=2, nrows=2, figsize=(12,8))
i = 0
for model_name, features in features_to_test.items():
col = i % 2
row = int(floor(i/2))
_train_X = train_X[:, features]
_test_X = test_X[:, features]
enet = ElasticNetCV(l1_ratio=np.array([.1, .5, .7, .9, .95, .99, 1]),
n_alphas=1000,
normalize=True,
selection='random',
random_state=1351)
enet.fit(_train_X, train_y)
coefs = pd.DataFrame({'Feature': features,
'Importance': np.abs(enet.coef_)})
coefs = coefs.set_index('Feature')\
.sort_values('Importance', ascending=False)
plot = coefs.plot(kind='bar', title='Typical feature importance chart', ax=axarr[(row,col)])
title = 'Features:' + model_name + r'; $R^2$ =' + \
str(round(enet.score(_test_X,test_y),3))
axarr[(row,col)].set_title(title)
i += 1
plt.tight_layout()
- What can we learn from this example?
First example/introducing the issues (continued)¶
- Here was the setup:
X,y = make_regression(n_samples=1000, n_features=10, n_informative=3, random_state=1234, shuffle=False, noise=10) X = np.append(X, X[:,[0]], axis=1)
- Some things to keep in mind:
- Interactions and correlated/dependent features makes intepretation tricky.
- Feature importance $\ne$ dependence of target on feature
- Feature importance $\approx$ contribution of feature to prediction function $f$
- This doesn't imply there do not exist other functions $f'$ where features that are 'unimportant' in $f$ are 'important'
Where we're headed:¶
- With this in mind, we're going to look at a number of methods for measuring feature importance.
- We can devide these methods into two basic classes:
- Algorithmic/prediction-function-specific methods: These exploit the structure of the learning algorithm/prediction function to construct some measure of the relative contribution of each feature.
- Model agnostic methods: These don't make assumptions on the algorithm or structure of the prediction function; can be applied to any black-box predictor.
Prediction-function-specific: Trees¶
- Our first algorithm/prediction function specific methods will address decision trees.
- We'll use the Iris Dataset, which is a canonical classification dataset (first used by R.A. Fisher) with:
- $\mathcal{Y} = $ {Setosa, Versicolour, Virginica} (three varieties of irises).
- $X = $ [sepal length, sepal width, petal length, petal width] $\in \mathbb{R}^4$
Prediction-function-specific: Trees (Continued)¶
- Let's consider a shallow tree built on this data.
hide_code_in_slideshow()
from sklearn.datasets import load_iris
from sklearn import tree
import graphviz
iris = load_iris()
clf = tree.DecisionTreeClassifier(max_depth=3)
clf = clf.fit(iris.data, iris.target)
dot_data = tree.export_graphviz(clf, out_file=None,
feature_names=iris.feature_names,
class_names=iris.target_names,
filled=True, rounded=True,
special_characters=True)
graph = graphviz.Source(dot_data)
display.display(graph)
- Question: Which of the feature ([sepal length, sepal width, petal length, petal width]) are most important? Why?
Prediction-function-specific: Trees (Continued)¶
Tree buiding reminders¶
- Consider node $t$ in a decision tree built on $N$ training data instances.
- Let node $t$ have $N_t$ node samples.
- We find the split $s_t$ at node $t$ such that $t_L$ and $t_R$ maximizes the decrease $$\Delta i(s, t) = i(t) − p_Li(t_L) − p_Ri(t_R)$$ where $p_L$ and $p_R$ are the probabilities an instance splits left and right, respectively, and $i(t)$ is some some impurity measure.
Prediction-function-specific: Trees (Continued)¶
Mean decrease impurity¶
- The mean decrease impurity $imp(X_m)$ for feature $X_m$ is: $$imp(X_m) = \sum_{v(s_t)=X_m}p(t)\Delta i(s_t,t)$$ Note $p(t) = N_t/N$ is the proportion of samples reaching node $t$ and $v(s_t)$ is the variable used in split $s_t$.
Prediction-function-specific: Trees (Continued)¶
$$imp(X_m) = \sum_{v(s_t)=X_m}p(t)\Delta i(s_t,t)$$
- Question: When will a feature be considered more important under this metric?
- Question: Why is this an algorithm/prediction-function specific method?
Prediction-function-specific: Trees (Continued)¶
- Implementation:
from collections import defaultdict
import numpy as np
def feature_importance_single_tree(tree, feature_names=iris.feature_names):
# Returns normed feature importance for a single sklearn.tree
# Note sklearn trees can handle instance weights, so we need
# to use weighted_n_node_samples
total_samples = np.sum(tree.weighted_n_node_samples)
feature_importance = defaultdict(float)
# Identify leaves as described in sklearn's plot_unveil_tree_structure.html
is_leaf = (tree.children_right == tree.children_left)
for ix in range(len(is_leaf)):
if not is_leaf[ix]:
impurity = tree.impurity[ix]
split_feature = tree.feature[ix]
num_at_node = tree.weighted_n_node_samples[ix]
# Get left child contribution
left_child = tree.children_left[ix]
left_decrease = tree.weighted_n_node_samples[left_child]/num_at_node * \
tree.impurity[left_child]
# Get right child contribution
right_child = tree.children_right[ix]
right_decrease = tree.weighted_n_node_samples[right_child]/num_at_node * \
tree.impurity[right_child]
delta = impurity - left_decrease - right_decrease
feature_importance[feature_names[split_feature]] \
+= num_at_node / total_samples * delta
norm = np.sum(feature_importance.values())
feature_importance = {key: val/norm for key, val in feature_importance.items()}
return feature_importance
Prediction-function-specific: Trees (Continued)¶
- Implementation vs. sklearn
imp = feature_importance_single_tree(clf.tree_)
display.display(pd.Series(imp).rename('Importance').to_frame()\
.reset_index().rename(columns={'index':'Feature'}))
| Feature | Importance | |
|---|---|---|
| 0 | petal length (cm) | 0.585616 |
| 1 | petal width (cm) | 0.414384 |
display.display(pd.DataFrame({'Feature': iris.feature_names,
'Importance':clf.feature_importances_})\
.sort_values('Importance', ascending=False))
| Feature | Importance | |
|---|---|---|
| 2 | petal length (cm) | 0.585616 |
| 3 | petal width (cm) | 0.414384 |
| 0 | sepal length (cm) | 0.000000 |
| 1 | sepal width (cm) | 0.000000 |
Prediction-function-specific: Trees (Continued)¶
- Question: How do we extend this to ensembles of trees?
- Answer: (Weighted) averages.
- Ex: Random Forests with $N_T$ trees:
$$Imp(X_m) = \frac{1}{N_T}\sum_T\sum_{t \in T:v(s_t)=X_m}p(t)\Delta i(s_t,t)$$
Prediction-function-specific: Trees (Continued)¶
- Implementation vs. sklearn
from sklearn.ensemble import RandomForestClassifier
forest = RandomForestClassifier(n_estimators=100) # Not tuned - just example
forest.fit(iris.data, iris.target)
display.display(pd.DataFrame({'Feature': iris.feature_names,
'Importance':forest.feature_importances_})\
.sort_values('Importance', ascending=False))
| Feature | Importance | |
|---|---|---|
| 2 | petal length (cm) | 0.449774 |
| 3 | petal width (cm) | 0.433002 |
| 0 | sepal length (cm) | 0.098417 |
| 1 | sepal width (cm) | 0.018807 |
forest_importance = defaultdict(float)
for tree in forest.estimators_:
tree_importance = feature_importance_single_tree(tree.tree_)
for key, val in tree_importance.items():
forest_importance[key] += val
forest_importance = {key:val/len(forest.estimators_)\
for key, val in forest_importance.items()}
display.display(pd.Series(forest_importance).rename('Importance').to_frame()\
.reset_index().rename(columns={'index':'Feature'})\
.sort_values('Importance', ascending=False))
| Feature | Importance | |
|---|---|---|
| 0 | petal length (cm) | 0.449774 |
| 1 | petal width (cm) | 0.433002 |
| 2 | sepal length (cm) | 0.098417 |
| 3 | sepal width (cm) | 0.018807 |
Prediction-function-specific: Trees (Continued)¶
- Let's think about potential variants/extensions to mean decrease impurity.
- How could we handle surrogate splits?
- Commercial implementation of CART adds contributions of surrogate splits:
- $\Delta i(s_t,t)$ for primary split variable
- $p^n \Delta i(s_t,t)$ for the $n$'th surrogate split variable (where $p \in [0,1]$ is a user-selected hyperparameter).
- Question: Can we think of any other variants or extensions?
- Consider collapsing the importance of dummy-encoded categoricals.
Prediction-function-specific: Linear models¶
- Question: How can we measure feature importance in a linear model?
$$imp(X_m) = \big\vert w_m \big\vert$$
- Question: Per usual, how does this notion of importance interact with preprocessing?
Model-agnostic feature importance:¶
- We've discussed two methods for algorithm or prediction-function specific feature importance measures:
- Trees (and their ensembles): Mean Decrease Impurity
- Linear methods: Absolute weights
- Question: How could we determine the importance of input features for an arbitrary black box prediction function?
- Hint 1: Think about defining importance as by measuring how much each feature independently contributes to the performance of the learned model.
- Hint 2: Imagine you were tasked with constructing a synthetic dataset with an unimportant feature -- how could you construct it? Given this insight, what operation could be applied to a feature that would be expected to have (a) no impact on the test set score for unimportant features, and (b) decrease performance for important features?
Model-agnostic feature importance:¶
Permutation Importance¶
- Described by Breiman in the original Random Forests paper (using OOB sample).
We describe it using an arbitrary held-out test set:
- Learn prediction function $f$ on training data.
- Measure the performance of $f$ on the test set - call this $s_{0}$.
- For each feature $i$ in $[1,\cdots,D]$:
- Permute just this feature in the test set.
- Pass this permuted test set through $f$ to obtain a new score $s_{i}$.
- Use either $s_{0} - s_{i}$ (or some appropriate variant on this idea) as feature importance.
- Question: Why bother permuting the feature? Why not replace it with some noise, like $\mathcal{N}(0,1)$?
Model-agnostic feature importance:¶
Permutation Importance¶
Question: Is this any different than the following procedure? If so, how?
- Learn prediction function $f$ on training data.
- Measure the performance of $f$ on the test set - call this $s_{0}$.
- For each feature $i$ in $[1,\cdots,D]$:
- Drop this feature from both the training and the test set.
- Learn a new prediction function without this feature -- call this function $f_{\setminus i}$
- Measure the performance of $f_{\setminus i}$ on the test set to obtain a new score $s_{i}$.
- Use either $s_{0} - s_{i}$ as feature importance.
- Key difference: Permutation importance (plus $\big\vert w \big \vert$ and MDI) describe the importance of each feature to a particular prediction function $f$. We don't claim that a feature with low permutation importance in $f$ couldn't have high importance in some other $f'$.
Feature importance pitfalls:¶
- To conclude our discussion of feature importance, we're going to look at an example to understand potential pitfalls.
- We will compare:
- Permutation importance
- MDI for a gradient boosting regressor
- $\big\vert w \big\vert$ for an elastic net
Feature importance pitfalls: Problem set up¶
import numpy as np
size = 1000
x1 = np.random.uniform(-10,10,size)
z1 = np.random.uniform(-10,10,size)
x2 = z1 + np.random.normal(0,2,size)
x3 = z1 + np.random.normal(0,2,size)
x4 = z1 + np.random.normal(0,2,size)
def actual_function(x1,z1):
return -1./2.*x1 + z1 + np.random.normal(0,1,len(x1))
y = actual_function(x1,z1)
X = np.stack([x1,x2,x3,x4]).T
- Question: Which feature is most important (i.e. contributes most to the true conditional mean function)?
- Question: Why might we expect this problem to challenge our feature importance methods?
hide_code_in_slideshow()
corr = pd.DataFrame(X).corr()
corr.index = ['$x_{}$'.format(x) for x in range(4)]
corr.columns = ['$x_{}$'.format(x) for x in range(4)]
display.display(display.Markdown('## Correlation matrix:'))
display.display(corr)
Correlation matrix:¶
| $x_0$ | $x_1$ | $x_2$ | $x_3$ | |
|---|---|---|---|---|
| $x_0$ | 1.000000 | -0.008428 | -0.004025 | -0.018237 |
| $x_1$ | -0.008428 | 1.000000 | 0.900929 | 0.899367 |
| $x_2$ | -0.004025 | 0.900929 | 1.000000 | 0.897771 |
| $x_3$ | -0.018237 | 0.899367 | 0.897771 | 1.000000 |
Feature importance pitfalls: Prediction functions¶
from sklearn.model_selection import train_test_split
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.model_selection import GridSearchCV
train_X, test_X, train_y, test_y = train_test_split(X,y,test_size=0.2,
random_state=1345134)
reg = GradientBoostingRegressor(n_estimators=200,subsample=0.5,
random_state=3141)
grid = GridSearchCV(reg,
param_grid={
'max_leaf_nodes':[10,25,50],
'min_samples_leaf':[10,25,50]
}, n_jobs=5, cv=5)
grid = grid.fit(train_X, train_y)
imp = pd.DataFrame({'Feature': ['$x_{}$'.format(i) for i in range(1,5)],
'GBM Importance':grid.best_estimator_.feature_importances_})
Feature Importance: Permutation Importance implementation¶
import eli5
from eli5.sklearn import PermutationImportance
perm = PermutationImportance(grid)
perm.fit(test_X, test_y)
imp['GBM Permutation'] = perm.feature_importances_
Feature Importance: Elastic Net¶
from sklearn.linear_model import ElasticNetCV
enet = ElasticNetCV(l1_ratio=np.array([.1, .5, .7, .9, .95, .99, 1]),
n_alphas=500,
normalize=True,
selection='random',
cv=5,
random_state=1351)
enet.fit(train_X, train_y)
imp['Elastic Net $\\vert w \\vert$'] = np.abs(enet.coef_)
perm2 = PermutationImportance(enet)
perm2.fit(test_X, test_y)
imp['Elastic Net Permutation'] = perm2.feature_importances_
Feature Importance: Results¶
- Recall $E[y~\big\vert~x_1,z] = -\frac{1}{2}x_1+z$. However, we don't observe $z$ -- instead have 3 indepedent noisy observations $x_2,x_3,x_4$. Trying to estimate $E[y~\big\vert~x_1,\cdots,x_4]$
hide_code_in_slideshow()
display.display(imp.set_index('Feature'))
| GBM Importance | GBM Permutation | Elastic Net $\vert w \vert$ | Elastic Net Permutation | |
|---|---|---|---|---|
| Feature | ||||
| $x_1$ | 0.269611 | 0.376578 | 0.493210 | 0.371113 |
| $x_2$ | 0.252952 | 0.163739 | 0.270237 | 0.137522 |
| $x_3$ | 0.240531 | 0.263513 | 0.377563 | 0.257554 |
| $x_4$ | 0.236906 | 0.191071 | 0.307340 | 0.192642 |
Feature Importance: Results¶
- Recall $E[y~\big\vert~x_1,z] = -\frac{1}{2}x_1+z$. However, we don't observe $z$ -- instead have 3 indepedent noisy observations $x_2,x_3,x_4$. Trying to estimate $E[y~\big\vert~x_1,\cdots,x_4]$
hide_code_in_slideshow()
sns.set_context('notebook',font_scale=2)
fig, ax = plt.subplots(figsize=(12,8))
plot = imp.set_index('Feature').plot(kind='bar', ax=ax)
- What are the implications, and what can we learn from this experiment?
Partial Dependence Plots¶
- Imagine we have a subset of features we think are important.
- This could be based on "feature importance" scores or coefficients, or other reasons (i.e. prior knowledge/research questions/etc.).
- We may want to dig deeper to explain the relationship between our predictions and these features.
- Question: Why? If we have MDI feature importance scores in hand, what do we not know?
- Directionality:
- Note in reality we probably have complex, non-monotonic relationships
- However even if we have a monotonic relationship, or even linear, our MDI wouldn't give any indication on the direction -- obviously weights in a linear model would.
- Partial dependence plots let us dig deeper and visualize the dependence of our prediction function (i.e. predict/predict_proba) on one or two of these features.
Partial Dependence Plots: Setup¶
- Consider an arbitrary prediction function $f$ learned over a training set $\mathcal{D}$.
This dataset includes $N$ observations $y_i$ of a target $y$ for $i = 1,2,\cdots,N$, along with $p$ features denoted $x_{i,j}$ for $j=1,2,\cdots,p$ and $i=1,2,\cdots,N$. This model generates predictions of the form: $$\hat{y}_i = f(x_{i,1},x_{i,2},\cdots,x_{i,p})$$
In the case of a single feature $x_j$, Friedman's partial dependence plots are obtained by computing the following average and plotting it over a useful range of $x$ values: $$\phi_j(x) = \frac{1}{N}\sum_{i=1}^N f(x_{i,1},\cdots,x_{i,j-1},x,x_{i,j+1},\cdots,x_{i,p})$$
- PDPs for more than one variable are computed (and plotted) similarly.
- Question: What would $\phi_j(x)$ be for a linear prediction function?
Partial Dependence Plots: Setup¶
$$\phi_j(x) = \frac{1}{N}\sum_{i=1}^N f(x_{i,1},\cdots,x_{i,j-1},x,x_{i,j+1},\cdots,x_{i,p})$$
- Question: Think about $\phi_j(x)$ -- what issues can we already anticipate?
- Imagine $P(x_j = x_{j+1}) = 1$. Then even though there is zero density where $x_j \ne x_{j+1}$, we still can evaluate $\phi_j(x)$ at $x \ne x_{i, j+1}$ for all $i \in {1,\cdots,n}$.
- Previewing what's next: Think about the potential impact of taking the average $\frac{1}{N}\sum_{i=1}^N (\cdot) $
PDP Example: California Housing¶
- Example derived from
sklearnPDP example, usingsklearnutilities. - While not shown, we first train a GBM on the California Housing dataset:
hide_code_in_slideshow()
print(cal_housing.DESCR)
California housing dataset.
The original database is available from StatLib
http://lib.stat.cmu.edu/datasets/
The data contains 20,640 observations on 9 variables.
This dataset contains the average house value as target variable
and the following input variables (features): average income,
housing average age, average rooms, average bedrooms, population,
average occupation, latitude, and longitude in that order.
References
----------
Pace, R. Kelley and Ronald Barry, Sparse Spatial Autoregressions,
Statistics and Probability Letters, 33 (1997) 291-297.
PDP Example: California Housing¶
- Example derived from
sklearnPDP example, usingsklearnutilities. - While not shown, we first train a GBM on the California Housing dataset:
hide_code_in_slideshow()
fig, ax = plt.subplots(figsize=(12,6))
plot = pd.Series(reg.feature_importances_,
index=names).rename('MDI').to_frame()\
.sort_values('MDI', ascending=False).plot(kind='bar',ax=ax,
title='Importance: Mean Decrease Impurity')
PDP Example: California Housing¶
- Next, we present the PDPs for several single features, and one pair of features.
hide_code_in_slideshow()
features = [0, 5, 1, 2, (5, 1)]
fig, ax = plt.subplots(figsize=(12,10))
fig, ax = plot_partial_dependence(reg, X_train, features,
feature_names=names,
n_jobs=3, grid_resolution=50,
ax=ax)
fig.suptitle('Partial dependence of house value on nonlocation features')
plt.subplots_adjust(top=0.9) # tight_layout causes overlap with suptitle
plt.show()
PDP Example: California Housing¶
- Note we don't plot the spatial features -- in fact, what would perhaps be most interesting (and a potential optional extension for indidivual exploration) would be to plot the long/lat PDP over a base map.
- An easy python first pass might simply use
geopandas.
- An easy python first pass might simply use
PDP Pitfalls:¶
- Consider the following PDP for a regression problem with $\mathcal{Y} = \mathbb{R}$ and $X = \mathbb{R}^3$.
hide_code_in_slideshow()
fig, ax = plt.subplots(figsize=(12,8))
plot_partial_dependence(grid.best_estimator_, Xs, [1],
n_jobs=1, grid_resolution=25, ax=ax)
plt.xlim(-1,1)
plt.ylim(-6,6)
plt.xlabel('$X_1$')
pass
- Question: What can we learn about the relationship between feature $X_2$ and our target $Y$?
PDP Pitfalls:¶
- Here was the data generating process:
size = 10000
X1 = np.random.uniform(-1,1,size)
X2 = np.random.uniform(-1,1,size)
X3 = np.random.uniform(-1,1,size)
eps = np.random.normal(0,0.5,size)
Y = 0.2*X1 - 5*X2 + 10*X2*np.where(X3>=0,1,0) + eps
Xs = np.stack([X1,X2,X3]).T
hide_code_in_slideshow()
fig, ax = plt.subplots(figsize=(12,8))
plt.scatter(X2[X3>=0], Y[X3>=0], color='blue', label='$X_3>=0$')
plt.scatter(X2[X3<0], Y[X3<0], color='green', label='$X_3<0$')
plt.ylabel('$Y$')
plt.xlabel('$X_2$')
plt.legend()
plt.show()
PDP Pitfalls:¶
- Recall $$y = 0.2 x_1 - 5 x_2 + 10 x_2 \mathbb{1}(x_3 >= 0) + \varepsilon$$
- Question: Why does the PDP fail to reveal the dependence of $y$ on $x_2$?
In his gradient boosting paper, Friedman identified this issue (here presented in our notation):
- In general, the functional form of $\phi_j(x)$ will depend on the particular values chosen for $\setminus j$. If, however, this dependence is not to strong than the average function can represent a useful summary of the partial dependence of $f$ on the chosen variable subset $j$.
PDP Pitfalls: Another option¶
- One response to this shortcoming has been development of "Individual Conditional Expectation" plots -- see Goldstein et al, 2014.
- Their insight was that taking an average is a post-processing step that can mask structure.
- We'll use the
SauceCat/PDPboximplementation.
hide_code_in_slideshow()
from pdpbox import pdp
train = pd.DataFrame(Xs)
train.columns = [str(x) for x in train.columns]
pdp_one = pdp.pdp_isolate(grid, train, '1')
pdp.pdp_plot(pdp_one, '$x_1$',
plot_org_pts=True, plot_lines=True,
center=False, frac_to_plot=1000)
- Question: Note the average (i.e. classic PDP plot) is shown in yellow. How do the additional traces aide clarify the dependence of $y$ on $x_1$?
Next steps¶
- We started by talking about feature importance.
- This gave a set of methods for trying to build insight into how much each feature contributed to the prediction function -- though we saw this is somewhat ill-defined. It didn't give insight into even the directionality of simple linear relationships -- let alone more complex stucture.
- Next we looked at PDPs
- PDPs give some insight into average relationship between one or two predictors and the target. However, this was shown to be potentially misleading, if the average relationship masks interactions.
- This led us to ICEplots, which plot a trace for each data instance separately.
- Note this trajectory (moving from global to data-instance-specific measures of importance) could be extended -- next steps might look at additive feature attribution methods such as SHAP values.